 |
 |
 |
Simple Atom Depth Index Calculator |
 |
 |
 |
1 Atoms exposition
The depth of atom from protein surface has been proposed as a criterion to
define protein structures. Current methods consist in evaluating the smaller
distance between an atom and a dot of the solvent accessible
surface [1] or the distance between an atom and its closest
solvent accessible neighbor [2]. These methods have the drawback
to lose the contribution from the three-dimensional molecular shape.
Figure 1:
Hen egg white lysozyme: 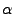 -carbon 47 (red) and 58 (green) are both on
the molecule surface, but accessibility of the former is quite
greater
-carbon 47 (red) and 58 (green) are both on
the molecule surface, but accessibility of the former is quite
greater
|
 |
The purpose of SADIC is to calculate atoms exposition keeping into account
the three-dimensional shape of the molecule. The program simulates the molecule
probing by a probe of given radius and calculates the exposed volume and
surface as seen by the probe, as well as the depth index.
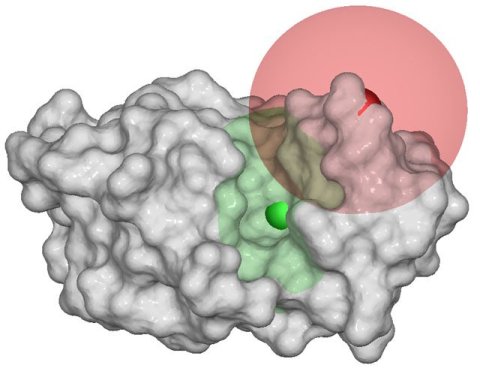
Figure:
Area E represents the space external to the molecule as
seen by the probe
|
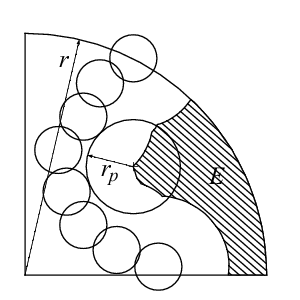 |
The depth index [3] is defined as
where 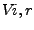 is the exposed volume of a sphere of radius r centered on atom
i and
is the exposed volume of a sphere of radius r centered on atom
i and 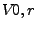 is the exposed volume of the same sphere when centered
on an isolated atom. Given this definition, the depth index results in a number
ranging from 0 to 2: its value is 0 if the atom is completely buried inside the
molecule and no external point is closer than r; a theoretical value of
2 would be obtained if the atom were completely isolated. If an atom center
lied on a theoretical perfect plane -- splitting the space into an
``internal'' and an ``external'' half-space -- its
is the exposed volume of the same sphere when centered
on an isolated atom. Given this definition, the depth index results in a number
ranging from 0 to 2: its value is 0 if the atom is completely buried inside the
molecule and no external point is closer than r; a theoretical value of
2 would be obtained if the atom were completely isolated. If an atom center
lied on a theoretical perfect plane -- splitting the space into an
``internal'' and an ``external'' half-space -- its 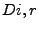 would be exactly
1.
would be exactly
1.
